Free Concurrency with GNU Parallel Parallelizing code without writing any
I am currently working on my thesis, which involves running simulations over and over again with varying parameters. GNU Parallel allows me to run these simulations concurrently with all possible combinations of parameters—without changing my codebase.
My simulation is implemented in Python. It takes some parameters as input, runs for some time, and finally prints the result as CSV to screen. Pretty simple.
Once I had set up the simulation and got it performing decently, I proceeded to the next step: running it with many different combinations of parameters in order to analyze and compare the results.
I was facing two problems:
-
How could I run the simulation for all possible combinations of parameters?
-
A single simulation takes about 1 minute to run. With a handful of arguments, each having many possible values, the number of parameter combinations explode, known as the curse of dimensionality. How can I take full advantage of the four cores on my computer—let alone the twenty-four cores at university—in order to reduce the total execution time to a fraction?
Going Down the Rabbit Hole
Addressing the first problem, i.e. calculating the combinations of all parameters, is straightforward. For n parameters I simply need n nested for-loops:
import simulation
for day in range(1, 31):
for algo in ['greedy', 'perfect', 'smart']:
for factor in [1, 5, 10]:
print(simulation.run(day, algo, factor))
That was easy. Granted, if I need to change the parameters I’d need to edit this file. I can’t do it on a case-by-case basis.
Not so fast, fella! What’s up with that print? How will I be able to
distinguish the different runs? Turns out I can no longer simply print the
result of each simulation to standard output as this would mingle all results.
And with no output I can no longer redirect the result to a file as I was doing
for a single simulation:
$ python simulation.py > result.csv
I now need to take care of saving each simulation’s result to a file in the Python script. Easy enough:
import simulation
for day in range(1, 31):
for algo in ['greedy', 'perfect', 'smart']:
for factor in [1, 5, 10]:
filename = 'output/{}-{}-{}.csv'.format(day, algo, factor)
with open(filename, 'w') as f:
f.write(simulation.run(day, algo, factor))
Noisy, but so what… Next up: reduce execution time by running simulations in parallel.
I’ve never worked with threads in Python. All I know is that Python—more precisely the CPython implementation—features a global interpreter lock, short GIL. The GIL allows exactly one native thread to execute at a time. In other words, because my problem is CPU bound, I don’t achieve any parallelization. (Note that, if your application is IO bound, threads can still give you a big performance boost since one thread can run while another thread is doing IO.)
There are definitely ways around the GIL, such as forking, using a different Python implementation, and many more according to the Python Wiki entry Parallel Processing and Multiprocessing in Python. But frankly, the options all sound like overkill, not to mention overwhelming. There must be an easier way to simply use all my CPU cores.
Introducing GNU Parallel
In short, GNU Parallel is a command line tool that allows you to run shell scripts in parallel, using all available CPU cores. It has many features, such as
- reading input from stdin,
- distributing jobs to remote machines over SSH, and
- options to control the number of cores used and the CPU usage.
Here’s a simple example taking input from stdin. We’ll use seq 5 to generate
the sequence of numbers from 1 to 5, each number on its own line, and pipe that
to parallel:
$ seq 5 | parallel echo
parallel will execute echo for each input line, passing the line as argument
to echo. It run as many processes simultaneously as there are number of cores.
The output looks as follows. Note that, due to the parallel execution, the order
may be different.
1
2
3
4
5
An alternative way of passing data to parallel is through the use of input
sources. The following example has the equivalent outcome as the one above:
$ parallel echo ::: $(seq 5)
Now the really cool part is that you can pass arbitrarily many input sources and
parallel will execute the command once for each item of the Cartesian product
of the input sources:
$ parallel echo ::: $(seq 5) ::: foo bar
This will print all combinations of the numbers 1 to 5 and the words foo and bar to screen.
1 foo
1 bar
2 foo
2 bar
3 foo
3 bar
4 foo
4 bar
5 foo
5 bar
If you want to run a snippet that is a bit more involved, pass it as a string and access the parameters individually:
$ parallel 'xyz -n{1} --word={2} > {2}-{1}.txt' ::: $(seq 10) ::: foo bar
As you can see, GNU Parallel addresses exactly the two problems I was facing: the
combination of all parameters and the parallel execution. Let’s see how we can
use parallel with the Python script above.
Running the Simulations with GNU Parallel
Remember how I hard coded the different parameters and combined them in the nested for-loops? Instead of doing that, we’ll have the arguments passed through the command line. All we have to do is adapt our Python script to read these input arguments. Thankfully, Python has the superb argparse module that makes it simple to build a nice command line interface.
Since we’ll be running just one simulation per script invocation, we can still simply print the results to stdout and use redirection to save it to a file.
With these two things in mind, this is how the script looks like:
import argparse
import simulation
parser = argparse.ArgumentParser(description='Run the simulation.')
parser.add_argument('DAY', type=int, help='The day number.')
parser.add_argument('ALGO', type=str, help='Which algorithm to use.')
parser.add_argument('FACTOR', type=int, help='Effect multiplication factor.')
args = parser.parse_args()
print(simulation.run(args.DAY, args.ALGO, args.FACTOR))
As you can see, the simulation script now has a simple interface, adhering to the UNIX philosophy Do One Thing and Do It Well. It runs one simulation for the specified parameters and outputs the results to stdout, allowing you to redirect it to a file, pipe it to another program, or simply look at it.
Now we’re ready to invoke the script with parallel and all the different
parameters:
$ parallel 'python simulation.py {1} {2} {3} > output/{1}-{2}-{3}.csv' ::: \
> $(seq 30) ::: \
> greedy perfect smart ::: \
> 1 5 10
This will run the snippet 30 × 3 × 3 = 270 times with varying arguments on all CPU cores.
Bonus: Keeping Track of Progress
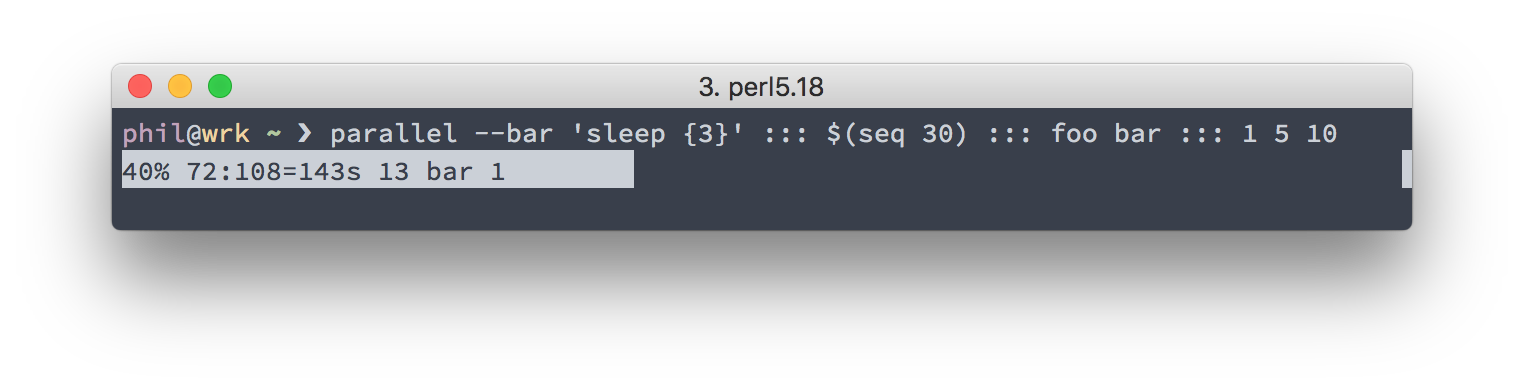
When running long tasks or a big number of tasks, it’s nice to know the progress
of the parallel invocation. Luckily, parallel accepts the --bar flag,
which gives you a nice-looking progress bar, showing you the percentage of
processed jobs and some further details such as the number of processed and
outstanding jobs.
Conclusion
By making sure that our script has a simple, yet functional interface and by printing to standard output, we were able to run it with GNU Parallel, achieving the desired goals of running the simulation with all possible combinations of parameters and making use of all CPU cores.
I’ve only scratched the surface of GNU Parallel’s possibilities. I hope this inspires you to include it in your daily workflow or when the occasional need for some quick and dirty concurrency arises. I encourage you to check out the excellent GNU Parallel tutorial, featuring dozens of examples. As it says on that tutorial:
Spend an hour walking through the tutorial. Your command line will love you for it.